| The
POOL Lab Research People Publications Genomes |
| Genetic Basis of Adaptive Evolution As further elaborated here, Drosophila melanogaster provides us with a remarkably efficient model system for investigating the genes and mutations that underlie adaptive differences between populations from contrasting environments. Understanding how adaptation operates at the molecular level is a fundamental concern for evolutionary biology, with several important questions not yet well understood, such as: * How polygenic is adaptation, and how many mutations underlie beneficial haplotypes at causative loci? * What functional categories of genes are involved, and are the causative mutations protein-coding or regulatory? * Do adaptive alleles arise from new mutations, or from standing genetic variation after an environmental change? * How geographically variable are adaptive genetic responses to parallel selective pressures? We use a variety of approaches to identify the genes and mutations underlying adaptive evolution, including: * Test for natural selection based on whole genome sequence variation. * RNA sequencing to quantify patterns of gene expression and alternative splicing. * Genome sequencing of offspring from genetic mapping crosses. * Molecular and transgenic studies to confirm the functional effects of selected variants. One example of using the Drosophila genetic toolkit to identify adaptive mutations comes from recent studies on pigmentation. I had previously documented striking melanic phenotypes among highland African populations of D. melanogaster and identified ebony as a gene of major effect in one melanic population (Pool and Aquadro 2007). 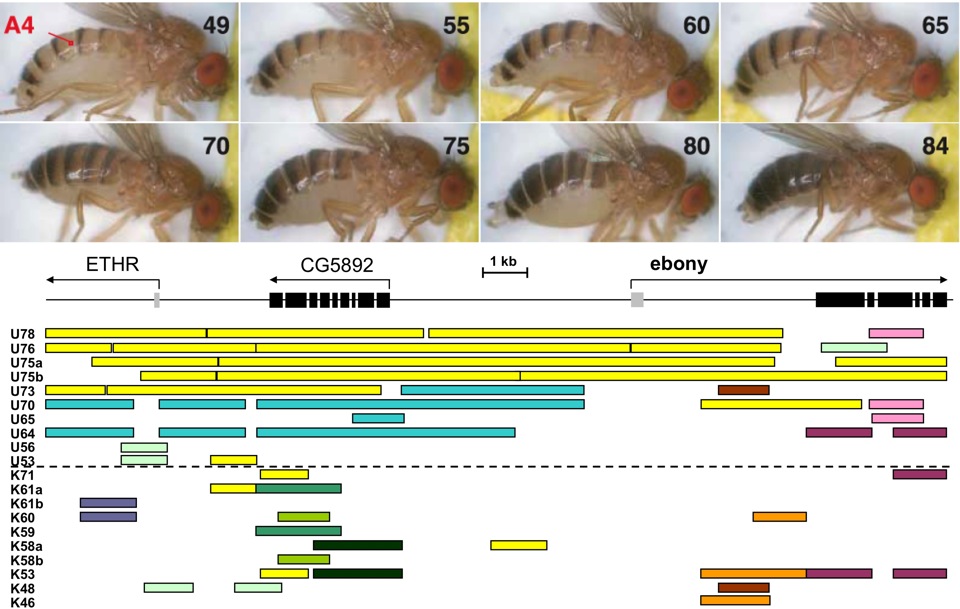 Top: Diversity of Top: Diversity of female abdominal pigmentation in African D. melanogaster. Bottom: Long identical haplotypes indicate recent natural selection at the ebony gene's upstream regulatory region in Uganda (U) but not Kenya (K). Pigmentation phenotypes (see numbers at left) are strongly correlated with ebony alleles in Uganda. I then collaborated with Mark Rebeiz and Sean Carroll in a fusion of molecular and population genetic approaches, with Mark ultimately identifying five specific mutations that conferred melanic pigmentation by altering the expression of ebony (Rebeiz et al. 2009). Part of our lab’s ongoing research is to combine population genomic data with genetic mapping approaches to identify causative pigmentation genes in the same population (where at least one causative locus more exists) and in other melanic populations. Based on preliminary data, the genetic basis of melanic pigmentation appears distinct in each African mountain range, potentially allowing us to investigate parallel instances of melanic evolution. We are open to studying any phenotypic difference (morphological, physiological, or behavioral) between D. melanogaster populations at the genetic level. Beyond pigmentation, we're also studying body size and cold tolerance. D. melanogaster originated from warm areas of sub-Saharan Africa, and our data indicate that the species adapted to cool climates not only as it expanded beyond Africa into Eurasia, but also twice within Africa (in Ethiopia and South Africa). We are combining population genomics, genetic mapping, and RNAseq data (gene expression and splicing) to investigated the genetic basis of this species' parallel evolution of cold tolerance. We're also studying body size evolution, especially for our Ethiopian flies, which are much larger than any previously documented population. Initial population genomic analysis has revealed some tantalizing candidate genes that have high genetic differentiation in this highland population. We're also interested in the cellular mechanisms underlying size differences. We tend to think of D. melanogaster as a diploid organism, but most tissues have elevated ploidy, which serves to regulate cell size and thus tissue size. Some of our candidate genes are known to influence body size by regulating the ploidy-elevating endocycle (DNA replication without cell division). Genes that are also implicated by genetic mapping will be confirmed via transgenic experiments, and ultimately the phenotypic influence of specific mutations can be tested.  Size comparison between highland Ethiopian (top) and lowland African D. melanogaster females. Size comparison between highland Ethiopian (top) and lowland African D. melanogaster females. Ethiopian flies show larger total body size (left) and disproportionately larger wings (center). The density of trichomes (one hair per wing cell) indicates that Ethiopian flies larger wings are partly due to larger cell size, suggesting higher DNA content per cell. |
Research People Publications Why Study Population Genetics? |